The Researcher's Degrees of Freedom: Balancing Autonomy and Reproducibility
Freedom, yeah!
Freedom for the researchers to access knowledge!
Freedom for the researchers to work with integrity!
Freedom for the researchers to publish their work!
Freedom… yeah?
But researcher's degrees of freedom is not as straightforward as it may seem, especially when striving for standardization and inter- and intra-lab reproducibility. In this blog, we will explore the concept of researcher's degrees of freedom and its impact in metagenomics workflows.
So What are Researcher's Degrees of Freedom?
Researcher's degrees of freedom refers to the inherent level of autonomy and discretion researchers have in designing and conducting experiments, analyzing data, and interpreting results. A high degree of freedom means researchers have numerous options to choose from in terms of study design, methods, and tools, as well as how they are applied (i.e. picking analysis and QC parameters and thresholds). However, with this freedom comes the responsibility of making multiple decisions throughout the workflow, which can affect the reproducibility of the study.
On the other hand, a low degree of freedom can also negatively impact the research by limiting the creativity and innovation of researchers, as well as by slowing down the research process. When it comes to reproducibility of studies exploring causation and correlation effects of the microbiome on Human health, it's a complex topic that can cause headaches as we try to strike the perfect balance between scientific innovation and the need for reproducible findings.
The Impact on Metagenomics Workflows:
In metagenomics, where workflows are complex and multi-step procedures, researcher's degrees of freedom plays a crucial role. A high degree of researcher freedom can negatively impact reproducibility by making it difficult for other researchers to understand how the study was conducted and to replicate the study's results¹(Schloss et al. 2018). It is important to note that not all decisions negatively alter the data generation process. However, as rule of thumb, the higher the degrees of freedom a workflow offers, the higher the number of bias-prone variables. Additionally, unchecked decision-making in data analysis can introduce unintentional or intentional bias, leading to false-positive or -negative results and inaccurate conclusions² (Holman et al. 2015).
The Complexity of Metagenomics Workflows:
Metagenomics workflows involve various decisions and trade-offs. Yes, unfortunately that means a high researcher's degrees of freedom. For example, there are multiple significant degrees of freedom in DNA sequencing workflows which researchers must choose from ranging from the decision between shotgun sequencing and 16S rDNA/amplicon sequencing, study design to library preparation and data analysis.
In their study of biases introduced during library preparation and sequencing for metagenomic studies, Poulsen et al. (2022)³ described and quantified that both library preparation and sequencing platform choice can introduce biases that alter the observed microbial composition. They performed an in silico analysis of published metagenomes and found that library preparation kits from different vendors as well as short-read and long-read sequencing platforms each introduced unique taxonomic biases. This aligns with the findings from Sato et al. (2019)⁴, who demonstrated the importance of selecting appropriate library preparation kits according to the specific purposes and targets of sequencing, especially in metagenomic studies where a diverse range of microbial species with varying degrees of GC content are present. Another study by Nearing et al. (2021)⁵ also described and quantified multiple steps where significant bias can be introduced, from sample collection and storage to bioinformatics analysis. They highlighted areas such as different DNA extraction efficiencies between microbes, introduction of contaminant DNA, and effects of long-term sample storage.
These studies, shed light on how different approaches and protocols lead to very different results. Furthermore, researchers may be inclined to test multiple approaches on their dataset and selectively report only the most favorable results. However, this risks overfitting the analysis to the specific dataset at hand. Such selective reporting could therefore inflate expectations of replicability beyond what the true performance of the method warrants (Ullmann et al. 2023)⁶.
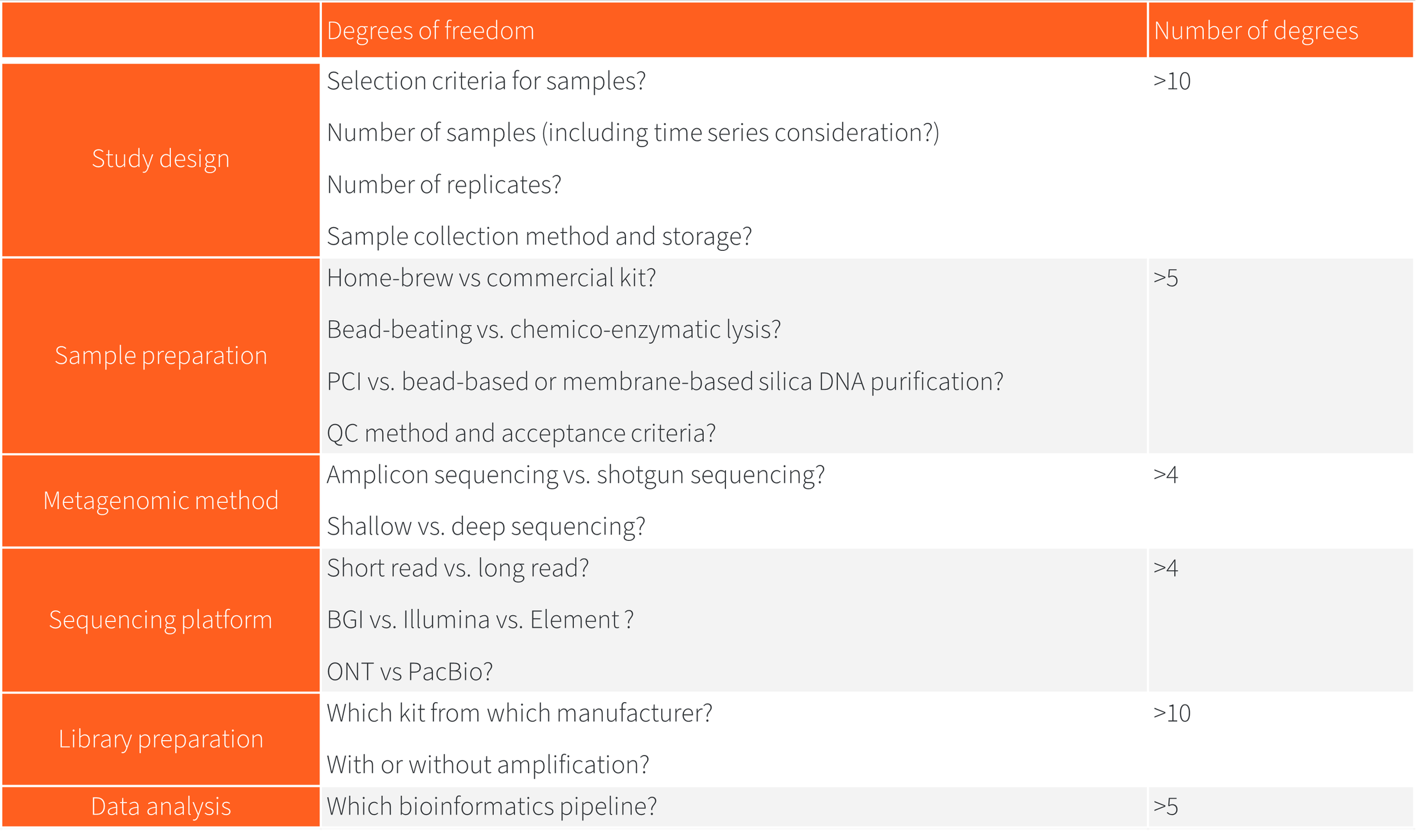
There are >40 significant degrees of freedom in DNA sequencing workflows:
Biases don’t just add, they multiply and accumulate along workflows, so that’s over 40,000+ possible combinations for each metagenomic project!
So, here's the thing… not all 40,000 combination would lead to same conclusion with the same confidence… or to the same conclusion at all!
Addressing Reproducibility Challenges:
To ensure reproducibility, it's essential to strike a balance between freedom and standardization. Providing clear and detailed descriptions of methods and tools used is crucial for other researchers to understand and replicate the study. Moreover, careful oversight and transparency in data analysis can minimize bias and improve the accuracy of conclusions.
But can we do proper metagenomics without sequencing?
Short answer: yes!
While sequencing has been a powerful tool for deep analysis of pure samples (i.e. from a single organism such as Human, animal, plant, isolated microbe, etc.), it has limitations and shortcomings when it comes to the analysis of complex samples containing a rich diversity of genetic materials, as in microbiome samples.
There is an alternative technology called optical mapping that offers a lower degree of freedom without compromising speed and performance. Optical mapping allows for the extraction of high quantities of genetic and taxonomic information directly from native metagenomic DNA, eliminating the need for amplification, library preparation, and sequencing.
Read this article to understand what makes optical mapping a paradigm shift in metagenomics, or download the DynaMAP white paper here.
Conclusion:
The researcher's degrees of freedom is a delicate balance between autonomy and reproducibility. While a high degree of freedom can foster innovation, it also poses challenges for reproducibility. It is crucial for researchers to provide detailed descriptions of methods and tools used and to exercise caution in data analysis. Additionally, exploring alternative technologies like optical mapping can offer a more straightforward approach to metagenomics analysis. By embracing the right balance, we can unlock the full potential of the microbiome and advance scientific knowledge.
References